Table of Contents
 |
| Python : Linear Regression |
Introduction to Linear Regression
Linear Regression is a supervised statistical technique where we try to estimate the dependent variable with a given set of independent variables. We assume the relationship to be linear and our dependent variable must be continuous in nature.In the following diagram we can see that as horsepower increases mileage decreases thus we can think to fit linear regression. The red line is the fitted line of regression and the points denote the actual observations.
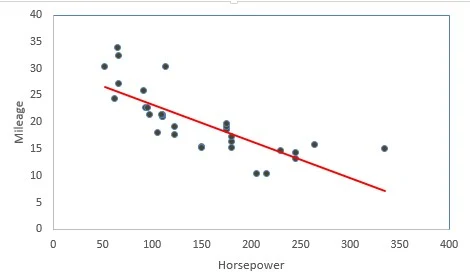
The vertical distance between the points and the fitted line (line of best fit) are called errors. The main idea is to fit this line of regression by minimizing the sum of squares of these errors. This is also known as principle of least squares.
Examples of Linear Regression
- Estimating the price (Y) of a house on the basis of its Area (X1), Number of bedrooms (X2), proximity to market (X3) etc.
- Estimating the mileage of a car (Y) on the basis of its displacement (X1), horsepower(X2), number of cylinders(X3), whether it is automatic or manual (X4) etc.
- To find the treatment cost or to predict the treatment cost on the basis of factors like age, weight, past medical history, or even if there are blood reports, we can use the information from the blood report.
Difference between Simple and Multiple Linear Regression
Simple Linear Regression Model: In this we try to predict the value of dependent variable (Y) with only one regressor or independent variable(X).The linear regression model:
Multiple Linear Regression Model: Here we try to predict the value of dependent variable (Y) with more than one regressor or independent variables.
Here 'y' is the dependent variable to be estimated, and X are the independent variables and ε is the error term.
 |
| Multiple Regression Equation |
Assumptions of linear regression
- There must be a linear relationship between the dependent and independent variables.
- Sample observations are independent.
- Error terms are normally distributed with mean 0.
- No multicollinearity - When the independent variables in my model are highly linearly related then such a situation is called multicollinearity.
- Error terms are identically and independently distributed. (Independence means absence of autocorrelation).
- Error terms have constant variance i.e. there is no heteroscedasticity.
- No outliers are present in the data.
Important Model Performance Metrics
Coefficient of Determination (R square)It suggests the proportion of variation in Y which can be explained with the independent variables. Mathematically, it is the ratio of predicted values and observed values, i.e.
 |
| RSquare |
If our fit is perfect then

If then R2 = 0 indicates a poor fit. Thus it lies between 0 and 1.
If the value of R2 is 0.912 then this suggests that 91.2% of the variation in Y can be explained with the help of given explanatory variables in that model. In other words, it explains the proportion of variation in the dependent variable that is explained by the independent variables.R square solely not such a good measure:
On addition of a new variable the error is sure to decrease, thus R square always increases whenever a new variable is added to our model. This may not describe the importance of a variable.
For eg. In a model determining the price of the house, suppose we had the variables GDP, Inflation rate, Area. If we add a new variable: no. of plane crashes (which is irrelevant) then still R square will increase.
Adjusted R square:
Adjusted R square is given by:
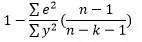 or
or
 |
| Adjusted R-Square |
where k is the no. of regressors or predictors.
Adjusted R square will always be less than or equal to R square.
On addition of a variable then R square in numerator and 'k' in the denominator will increase. If the variable is actually useful then R square will increase by a large amount and 'k' in the denominator will be increased by 1. Thus the magnitude of increase in R square will compensate for increase in 'k'. On the other hand, if a variable is irrelevant then on its addition R square will not increase much and hence eventually adjusted R square will increase.
Thus as a general thumb rule if adjusted R square increases when a new variable is added to the model, the variable should remain in the model. If the adjusted R square decreases when the new variable is added then the variable should not remain in the model.Why error terms should be normally distributed?
For parameter estimate (i.e. estimating the βi’s) we don't need that assumption. But, if it is not a normal distribution, some of those hypotheses tests which we will be doing as part of diagnostics may not be valid.
For example: To check whether the Beta (the regression coefficient) is significant or not, I'll do a T-test. So, if my error is not a normal distribution, then the statistic I derive may not be a T-distribution. So, my diagnostic test or hypotheses test is not valid. Similarly, F-test for linear regression which checks whether any of the independent variables in a multiple linear regression model are significant will be not be viable.Why is expectation of error always zero?
The error term is the deviation between observed points and the fitted line. The observed points will be above and below the fitted line, so if I took the average of all the deviations, it should be 0 or near 0. Zero conditional mean is there which says that there are both negative and positive errors which cancel out on an average. This helps us to estimate dependent variable precisely.
Why multicollinearity is a problem?
If my Xi’s are highly correlated then |X’X| will be close to 0 and hence inverse of (X’X) will not exist or will be indefinitely large. Mathematically, which will be indefinitely large in presence of multicollinearity. Long story in short, multicollinearity increases the estimate of standard error of regression coefficients which makes some variables statistically insignificant when they should be significant.
How can you detect multicollinearity?
- Bunch Map Analysis: By plotting scatter plots between various Xi’ s we can have a visual description of how the variables are related.
- Correlation Method: By calculating the correlation coefficients between the variables we can get to know about the extent of multicollinearity in the data.
- VIF (Variance Inflation Factor) Method: Firstly we fit a model with all the variables and then calculate the variance inflation factor (VIF) for each variable. VIF measures how much the variance of an estimated regression coefficient increases if your predictors are correlated. The higher the value of VIF for ith regressor, the more it is highly correlated to other variables.
Variance inflation factor (VIF) for an explanatory variable is given 1/(1-R^2 ) . Here, we take that particular X as response variable and all other explanatory variables as independent variables. So, we run a regression between one of those explanatory variables with remaining explanatory variables.Detecting heteroscedasticity
- Graphical Method: Firstly do the regression analysis and then plot the error terms against the predicted values( Yi^). If there is a definite pattern (like linear or quadratic or funnel shaped) obtained from the scatter plot then heteroscedasticity is present.
- Goldfeld Quandt (GQ)Test: It assumes that heteroscedastic variance σi2 is positively related to one of the explanatory variables And errors are assumed to be normal. Thus if heteroscedasticity is present then the variance would be high for large values of X.
Steps for GQ test:
- Order/ rank (ascending) the observations according to the value of Xi beginning with the lowest X value.
- Omit ‘c’ central observations and ide the remaining (n-c) observations into 2 groups of (n-c)/2 observations each.
- Fit separate OLS regression to both the groups and obtain residual sum of squares (RSS1 and RSS2) for both the groups.
- Obtain F = RSS2/ RSS1
It follows F with ((n-c)/2-k) d.f. both both numerator and denominator.About Dataset
Where k is the no. of parameters to be estimated including the intercept.
If errors are homoscedastic then the two variances RSS2 and RSS1 turn out to be equal i. e. F will tend to 1.
We have 1030 observations on 9 variables. We try to estimate the Complete compressive strength(CRS) using:
- Cement - kg in a m3 mixture
- Blast Furnace Slag - kg in a m3 mixture
- Fly Ash - kg in a m3 mixture
- Water - kg in a m3 mixture
- Superplasticizer - kg in a m3 mixture
- Coarse Aggregate - kg in a m3 mixture
- Fine Aggregate - kg in a m3 mixture
- Age - Day (1-365)
Dataset - Download Data
Python Code : Linear Regression
Importing libraries
Numpy, pandas and matplotlib.pyplot are imported with aliases np, pd and plt respectively.import numpy as np import pandas as pd import matplotlib.pyplot as pltLoading the data We load our data using pd.read_csv( )
data = pd.read_csv("Concrete_Data.csv")Now the data is ided into independent (x) and dependent variables (y)
x = data.iloc[:,0:8] y = data.iloc[:,8:]
Splitting data into training and test sets
Using sklearn we split 80% of our data into training set and rest in test set. Setting random_state will give the same training and test set everytime on running the code.from sklearn.cross_validation import train_test_split x_train,x_test,y_train,y_test = train_test_split(x,y,test_size = 0.2,random_state = 100)
Running linear regression using sklearn
Using sklearn linear regression can be carried out using LinearRegression( ) class. sklearn automatically adds an intercept term to our model.from sklearn.linear_model import LinearRegression lm = LinearRegression() lm = lm.fit(x_train,y_train) #lm.fit(input,output)The coefficients are given by:
lm.coef_
array([[ 0.12415357, 0.10366839, 0.093371 , -0.13429401, 0.28804259, 0.02065756, 0.02563037, 0.11461733]])To store coefficients in a data frame along with their respective independent variables -
coefficients = pd.concat([pd.DataFrame(x_train.columns),pd.DataFrame(np.transpose(lm.coef_))], axis = 1)
0 Cement 0.124154 1 Blast 0.103668 2 Fly Ash 0.093371 3 Water -0.134294 4 Superplasticizer 0.288043 5 CA 0.020658 6 FA 0.025630 7 Age 0.114617The intercept is:
lm.intercept_
array([-34.273527])To predict the values of y on the test set we use lm.predict( )
y_pred = lm.predict(x_test)
Errors are the difference between observed and predicted values.y_error = y_test - y_predR square can be obbtained using sklearn.metrics ( ):
from sklearn.metrics import r2_score r2_score(y_test,y_pred)
0.62252008774048395
Running linear regression using statsmodels
It is to be noted that statsmodels does not add intercept term automatically thus we need to create an intercept to our model.import statsmodels.api as sma X_train = sma.add_constant(x_train) ## let's add an intercept (beta_0) to our model X_test = sma.add_constant(x_test)Linear regression can be run by using sm.OLS:
import statsmodels.formula.api as sm lm2 = sm.OLS(y_train,X_train).fit()The summary of our model can be obtained via:
lm2.summary()
""" OLS Regression Results ============================================================================== Dep. Variable: CMS R-squared: 0.613 Model: OLS Adj. R-squared: 0.609 Method: Least Squares F-statistic: 161.0 Date: Wed, 03 Jan 2018 Prob (F-statistic): 4.37e-162 Time: 21:29:10 Log-Likelihood: -3090.4 No. Observations: 824 AIC: 6199. Df Residuals: 815 BIC: 6241. Df Model: 8 Covariance Type: nonrobust ==================================================================================== coef std err t P>|t| [0.025 0.975] ------------------------------------------------------------------------------------ const -34.2735 29.931 -1.145 0.253 -93.025 24.478 Cement 0.1242 0.010 13.054 0.000 0.105 0.143 Blast 0.1037 0.011 9.229 0.000 0.082 0.126 Fly Ash 0.0934 0.014 6.687 0.000 0.066 0.121 Water -0.1343 0.046 -2.947 0.003 -0.224 -0.045 Superplasticizer 0.2880 0.102 2.810 0.005 0.087 0.489 CA 0.0207 0.011 1.966 0.050 2.79e-05 0.041 FA 0.0256 0.012 2.131 0.033 0.002 0.049 Age 0.1146 0.006 19.064 0.000 0.103 0.126 ============================================================================== Omnibus: 3.757 Durbin-Watson: 2.033 Prob(Omnibus): 0.153 Jarque-Bera (JB): 3.762 Skew: -0.165 Prob(JB): 0.152 Kurtosis: 2.974 Cond. No. 1.07e+05 ============================================================================== Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified. [2] The condition number is large, 1.07e+05. This might indicate that there are strong multicollinearity or other numerical problems. """The predicted values for test set are given by:
y_pred2 = lm2.predict(X_test)
Note that both y_pred and y_pred2 are same. It's just these are calculated via different packages.Calculate R-Squared and Adjusted R-Squared Manually on Test data
We can also calculate r-squared and adjusted r-squared via formula without using any package.import numpy as np y_test = pd.to_numeric(y_test.CMS, errors='coerce') RSS = np.sum((y_pred2 - y_test)**2) y_mean = np.mean(y_test) TSS = np.sum((y_test - y_mean)**2) R2 = 1 - RSS/TSS R2 n=X_test.shape[0] p=X_test.shape[1] - 1 adj_rsquared = 1 - (1 - R2) * ((n - 1)/(n-p-1)) adj_rsquaredR-Squared : 0.6225
Adjusted RSquared : 0.60719
Detecting Outliers:
Firstly we try to get the studentized residuals using get_influence( ). The studentized residuals are saved in resid_student.
influence = lm2.get_influence() resid_student = influence.resid_studentized_externalCombining the training set and the residuals we have:
Cement Blast Fly Ash Water Superplasticizer CA FA Age \ 0 540.0 0.0 0.0 162.0 2.5 1040.0 676.0 28.0 1 540.0 0.0 0.0 162.0 2.5 1055.0 676.0 28.0 2 332.5 142.5 0.0 228.0 0.0 932.0 594.0 270.0 3 332.5 142.5 0.0 228.0 0.0 932.0 594.0 365.0 4 198.6 132.4 0.0 192.0 0.0 978.4 825.5 360.0 Studentized Residuals 0 1.559672 1 -0.917354 2 1.057443 3 0.637504 4 -1.170290
resid = pd.concat([x_train,pd.Series(resid_student,name = "Studentized Residuals")],axis = 1) resid.head()If the absolute value of studentized residuals is more than 3 then that observation is considered as an outlier and hence should be removed. We try to create a logical vector for the absolute studentized residuals more than 3
Cement Blast Fly Ash Water Superplasticizer CA FA Age \
649 166.8 250.2 0.0 203.5 0.0 975.6 692.6 3.0
Studentized Residuals
649 3.161183
resid.loc[np.absolute(resid["Studentized Residuals"]) > 3,:]The index of the outliers are given by ind:
ind = resid.loc[np.absolute(resid["Studentized Residuals"]) > 3,:].index ind
Int64Index([649], dtype='int64')Dropping Outlier
Using the drop( ) function we remove the outlier from our training sets!
y_train.drop(ind,axis = 0,inplace = True) x_train.drop(ind,axis = 0,inplace = True) #Interept column is not there X_train.drop(ind,axis = 0,inplace = True) #Intercept column is thereDetecting and Removing Multicollinearity
We use the statsmodels library to calculate VIF
from statsmodels.stats.outliers_influence import variance_inflation_factor [variance_inflation_factor(x_train.values, j) for j in range(x_train.shape[1])]
[15.477582601956859, 3.2696650121931814, 4.1293255012993439, 82.210084751631086, 5.21853674386234, 85.866945489015535, 71.816336942930675, 1.6861600968467656]We create a function to remove the collinear variables. We choose a threshold of 5 which means if VIF is more than 5 for a particular variable then that variable will be removed.
def calculate_vif(x): thresh = 5.0 output = pd.DataFrame() k = x.shape[1] vif = [variance_inflation_factor(x.values, j) for j in range(x.shape[1])] for i in range(1,k): print("Iteration no.") print(i) print(vif) a = np.argmax(vif) print("Max VIF is for variable no.:") print(a) if vif[a] <= thresh : break if i == 1 : output = x.drop(x.columns[a], axis = 1) vif = [variance_inflation_factor(output.values, j) for j in range(output.shape[1])] elif i > 1 : output = output.drop(output.columns[a],axis = 1) vif = [variance_inflation_factor(output.values, j) for j in range(output.shape[1])] return(output) train_out = calculate_vif(x_train)Now we view the training set
train_out.head()
Cement Blast Fly Ash Superplasticizer Age
337 275.1 0.0 121.4 9.9 56 384 516.0 0.0 0.0 8.2 28 805 393.0 0.0 0.0 0.0 90 682 183.9 122.6 0.0 0.0 28 329 246.8 0.0 125.1 12.0 3Removing the variables from the test set.
x_test.head() x_test.drop(["Water","CA","FA"],axis = 1,inplace = True) x_test.head()
Cement Blast Fly Ash Superplasticizer Age 173 318.8 212.5 0.0 14.3 91 134 362.6 189.0 0.0 11.6 28 822 322.0 0.0 0.0 0.0 28 264 212.0 0.0 124.8 7.8 3 479 446.0 24.0 79.0 11.6 7Running linear regression again on our new training set (without multicollinearity)
import statsmodels.api as sma import statsmodels.formula.api as sm train_out = sma.add_constant(train_out) ## let's add an intercept (beta_0) to our model x_test.drop(["Water","CA","FA"],axis = 1,inplace = True) X_test = sma.add_constant(x_test) lm2 = sm.OLS(y_train,train_out).fit() lm2.summary()
""" OLS Regression Results ============================================================================== Dep. Variable: CMS R-squared: 0.570 Model: OLS Adj. R-squared: 0.567 Method: Least Squares F-statistic: 216.3 Date: Wed, 10 Jan 2018 Prob (F-statistic): 6.88e-147 Time: 15:14:59 Log-Likelihood: -3128.8 No. Observations: 823 AIC: 6270. Df Residuals: 817 BIC: 6298. Df Model: 5 Covariance Type: nonrobust ==================================================================================== coef std err t P>|t| [0.025 0.975] ------------------------------------------------------------------------------------ const -11.1119 1.915 -5.803 0.000 -14.871 -7.353 Cement 0.1031 0.005 20.941 0.000 0.093 0.113 Blast 0.0721 0.006 12.622 0.000 0.061 0.083 Fly Ash 0.0614 0.009 6.749 0.000 0.044 0.079 Superplasticizer 0.7519 0.077 9.739 0.000 0.600 0.903 Age 0.1021 0.006 16.582 0.000 0.090 0.114 ============================================================================== Omnibus: 0.870 Durbin-Watson: 2.090 Prob(Omnibus): 0.647 Jarque-Bera (JB): 0.945 Skew: 0.039 Prob(JB): 0.623 Kurtosis: 2.853 Cond. No. 1.59e+03 ==============================================================================Checking normality of residuals We use Shapiro Wilk test from scipy library to check the normality of residuals.
- Null Hypothesis: The residuals are normally distributed.
- Alternative Hypothesis: The residuals are not normally distributed.
from scipy import stats stats.shapiro(lm2.resid)
(0.9983407258987427, 0.6269884705543518)Since the pvalue is 0.6269 thus at 5% level of significance we can say that the residuals are normally distributed.
Checking for autocorrelation To ensure the absence of autocorrelation we use Ljungbox test.
- Null Hypothesis: Autocorrelation is absent.
- Alternative Hypothesis: Autocorrelation is present.
from statsmodels.stats import diagnostic as diag diag.acorr_ljungbox(lm2.resid , lags = 1)
(array([ 1.97177212]), array([ 0.16025989]))Since p-value is 0.1602 thus we can accept the null hypothesis and can say that autocorrelation is absent.
Checking heteroscedasticity Using Goldfeld Quandt we test for heteroscedasticity.
- Null Hypothesis: Error terms are homoscedastic
- Alternative Hypothesis: Error terms are heteroscedastic.
import statsmodels.stats.api as sms from statsmodels.compat import lzip name = ['F statistic', 'p-value'] test = sms.het_goldfeldquandt(lm2.resid, lm2.model.exog) lzip(name, test)[('F statistic', 0.9903), ('p-value', 0.539)]
The p-value is 0.539 hence we can say that the residuals have constant variance. Hence we can say that all the assumptions of our linear regression model are satisfied.


How to download the data used in this article?
ReplyDeleteThe download link has been added. Thanks!
Delete
ReplyDeleteI am new bee to R and Python world. So I just want to know whether
Data Science and R course includes Python and R both?.
I am in search of course where I want to learn both languages.
Please let me know if you have any course for it along with tution fee
and batch starts date.
I have replied back on your email address. Thanks!
DeleteVery nice article, thank you.
ReplyDeleteI tried to apply your formulas on the data, but I noticed that after removing multicollinearity columns then I tried OLS again, multicollinearit didn't remove. Did you notice?
ReplyDeleteHi, I hope this code is having some error "from sklearn.cross_validation import train_test_split".
ReplyDeletetrain_test_split feature is available in sklearn.model_selection.
thank you very much
ReplyDeletenice article
ReplyDeleteNice explanation. Please let me know how we learn machine learning concepts from scratch?
ReplyDelete